In this article we will discuss about:- 1. Meaning of Chromosome 2. Morphology of Chromosome 3. Structure 4. Autosome and Sex Chromosome 5. Chemical Nature 6. Molecular Organisation 7. Special Types.
Contents:
- Meaning of Chromosome
- Morphology of Chromosome
- Structure of Chromosome
- Autosome and Sex Chromosome
- Chemical Nature of the Eukaryotic Chromosomes
- Molecular Organisation of Chromatin
- Special Types of Chromosomes
1. Meaning of Chromosome:
Chromosomes are thread-like structures, made up of the genetic material of the cell-DNA and protein; visible during the cell division under microscope, only after they are stained with dyes. Literally it means coloured body (chrom; colour, soma; body). These nuclear components are self-reproducing and play a vital role in heredity, mutation, variation and evolution.
ADVERTISEMENTS:
Actually, chromosomes are the structures that serve as the vehicle for transmission of genetic information. The present name chromosome was coined by W. Waldeyer (1888) to the darkly stained bodies of nucleus. Long before that, in 1842, Karl Nagli observed rod-like chromosomes in the nuclei of plant cells. Thereafter, many scientists have studied different aspects of both prokaryotic and eukaryotic chromosomes.
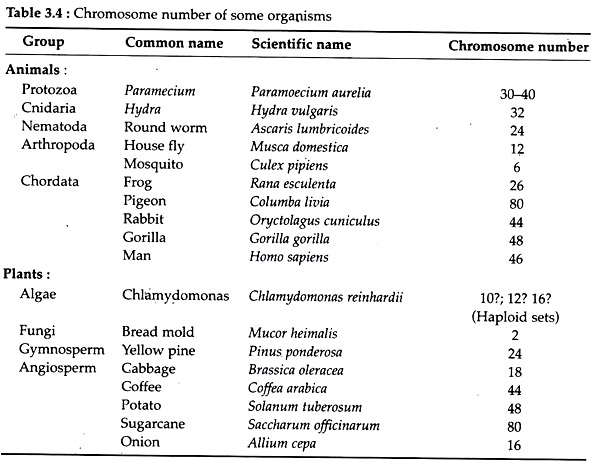
Number:
For a particular species, the number of chromosome is generally constant. The genome of prokaryotes are contained in single chromosome, in contrast, the genomes of eukaryotes are composed of multiple chromosomes. The number of chromosomes present in the gametic cell (e.g., sperms and ova) of any eukaryotic organism is called haploid set which is usually indicated by the word ‘n’ (or 1 x n = n).
The number of set of chromosomes present in the somatic or body cells of most organism is called diploid set and is indicated by ‘2n’. The number of chromosomes bears no direct relationship to the position of the species in the phylogenetic scheme of classification as evidenced from the Table 3.4. Man, the highly evolved animal has 46 chromosomes, while pigeon has 80 chromosomes.
2. Morphology of Chromosome:
The morphology of the chromosomes is best studied during meta- phase and anaphase stages of mitosis.
Size:
The size of chromosomes varies from one organism to another. Further, the chromosomes in a cell are never alike in size; some may be exceptionally large while others may be too small. The largest chromosomes are lamp brush chromosomes present in certain vertebrate oocytes. However, most metaphase chromosomes fall within a range of 3 µm in Drosoplrila, to 5 µm in man and 8 pm in maize.
ADVERTISEMENTS:
Shape:
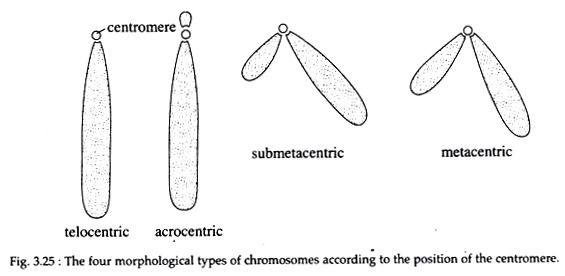
The shape of chromosomes is determined by the position of the centromere. Chromosomes are classified into four types according to their shape (Fig. 3.25).
(i) Telocentric:
The rod-like chromosomes having the centromere located on one end.
(ii) Acrocentric:
These are also rod-like chromosomes that have a very small or even imperceptible short arm.
(iii) Sub-metacentric:
The sub-metacentric chromosomes are ‘J’-or ‘L’-shaped having two arms of unequal length. The centromere is located away from the centre.
ADVERTISEMENTS:
(iv) Metacentric:
The ‘V’-shaped chromosomes having two equal or almost equal arms. The centromere occurs at the centre.
3. Structure of Chromosome:
Cytogenesis’s have introduced many terms for different parts of chromosomes that can be visualized in mitotic chromosomes.
These are as follows:
Chromatid:
At metaphase, each chromosome consists of two symmetrical rod- shaped structures, the chromatids, that are attached to each other firmly at the centromere and more loosely along their entire length. At the start of anaphase, the chromatids get separated and start to migrate to opposite poles. Each chromatid contains a single DNA molecule.
Chromonema:
During the early stage of condensation (prophase), the chromatids look like very thin filaments and are referred to as chromonemata. This chromonemata form the gene-bearing portion of the chromosomes.
Chromomeres:
During interphase, some bead like structures are found along the length of the chromosomes which are called chromomeres. These are actually accumulated chromatin materials and tightly folded regions of DNA. In polytene chromosomes, chromomeres are clearly visible where they become aligned side by side. At metaphase, the chromomere is so tightly coiled that chromomeres become invisible.
Centromere or Kinetochore:
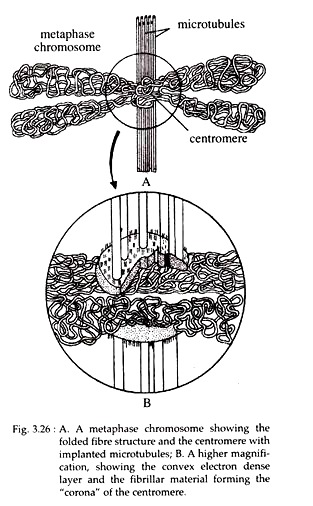
Each chromosome contains a site where the outer surfaces are markedly indented. This site is called centromere or primary constriction. It serves both as the attachment site for microtubules during cell division and as the site of association of sister chromatids.
Centromeres of higher eukaryotic cells are characterized by extensive regions of heterochromatins consisting of highly repetitive satellite DNA sequences. To the DNA sequences of centromere, some special proteins remain associated and form disc-shaped structure called kinetochore.
In electron microscope, it shows a trilaminar structure with a dense outer proteinaceous layer, a middle layer of low density, and a dense inner layer tightly bound to the centromere. The DNA of centromere does not exist in the form of nucleosome. Further, emanating from the convex surface of the outer layer, in addition to the microtubules, a “corona” or “collar” of fine filaments has been observed (Fig. 3.26).
During mitosis, 4 to 40 microtubules of mitotic spindle become attached to the kinetochore. Proteins associated with the kinetochore act as “molecular motors” that drive the movement of chromosomes along the spindle fibres, segregating the chromosomes during anaphase.
Actually, the kinetochore provides a center of assembly for microtubules, i.e., it serves as a nucleation centre to the polymerization of tubulin protein into microtubules. The chromosomes of most organisms contain only one centromere and are known as ‘mono-centric chromosome’. Some species have ‘diffused kinetochore, with microtubules attached along the length of the chromosome. These are called ‘holocentric chromosome’.
Sometimes a chromosome may break and fuse with another producing chromosome without kinetochore, referred as acentric chromosome or with two kinetochores, referred to as dicentric chromosome. However, both types are unstable.
Telomere:
The repeated sequences at the ends of the eukaryotic chromosomes are called telomeres that form a ‘cap’ at each end of the chromosome. They play some critical role in chromosome replication and maintenance. They protect the chromosomes from nuclease and other destabilizing influences and prevent the ends of chromosomes from fusing with one another.
Thus, telomeres facilitate interaction between the ends of the chromosomes and the nuclear envelopes in some type of cells. Normal cells are able to divide only a limited number of times before they show signs of ‘aging’, due to progressive shortening of the telomeres.
Recent studies have shown that a number of human cancers, possess a high telomerase activity (not present in the normal counterparts) which maintains the length of the telomeres from one generation to the next.
Secondary constriction:
Constrictions other than primary constriction, at any point of the chromosome are called secondary constrictions. These are constant in their position and are distinguished from the primary constriction by the absence of marked angular deviations of the chromosomal segments during anaphase.
Nucleolar Organizer:
Certain secondary constrictions contain the genes coding for 18S and 28S ribosomal RNA and induce the formation of nucleoli. These are referred to as nucleolar organizer. In human, the nucleolar organizers are located in the secondary constriction of chromosomes 13, 14, 15, 21 and 22, all of which are acrocentric and have satellite.
Satellite:
Sometimes the chromosomes bear a rounded knob-like body connected with the rest of the chromosomes by a thin chromatin filament. This structure is called ‘satellite’. Chromosomes bearing this satellite are designated as ‘Sat chromosomes’. The shape and size of satellite remain constant.
Karyotype and Idiogram:
A complete set of all the metaphase chromosomes in a cell is called its karyotype. The term, however, has been given to a group of characteristics that identifies a particular set of chromosomes; i.e., the number of chromosome, relative size, position of centromere, length of arms, secondary constrictions, and satellite. Karyotype is species specific.
A diagrammatic representation of the karyotype of a species is called ‘idiogram’. Generally, in an idiogram, the chromosomes of an organism are ordered according to the size and position of the centromere. Karyotypes of different species are sometimes compared and similarities in karyotypes are presumed to represent evolutionary relationship.
Some species may have special characteristics in their karyotypes, e.g., the mouse has acrocentric chromosomes, many amphibians have only metacentric chromosomes and plants frequently have heterochromatic regions at the telomeres.
4. Autosome and Sex Chromosome:
Chromosomes related to the sex of the organism are called sex chromosomes. Chromosomes other than sex chromosomes are called autosomes. In animals and in some plants there are differences in the chromosome complement of male and female cells.
One sex has a matched pair of sex chromosomes the other sex has an unmatched pair or a single sex chromosome. For example, human females have two X chromosomes (XX), while males have one X and one Y (XY) chromosome as their sex chromosomes. In human, both male and female have 44 autosomes.
5. Chemical Nature of the Eukaryotic Chromosomes:
Each eukaryotic chromosome consists of one linear, unbroken, double-stranded DNA molecule running throughout its length and contains about twice as much protein by weight as DNA. Each species has a characteristic content of DNA, which is constant in all the individuals of that species and has thus been called the C-value.’
The amount of genetic material in eukaryotic cell is nearly 100 times greater than that in prokaryotic cells. However, there is no direct relationship between the C value and the structural or organisational complexity of the organism. In eukaryotic cells, the large amount of DNA remains in very compact form in its nucleus.
The complexed structure between eukaryotic DNA and protein are called chromatin. The fundamental structure of chromatin is essentially identical in all eukaryotes. There are two types of chromatin – euchromatin and heterochromatin.
In 1928, Heitz defined heterochromatin as those regions of the chromosome that remain condensed during interphase to early prophase and that stains darkly. Euchromatin, on the other hand, is chromatin that stains lightly. It is uncoiled during interphase, but becomes condensed during mitosis. Most of the genome consists of euchromatin.
Functionally, euchromatin is genetically active i.e., it contains genes that are being expressed; whereas heterochromatin is genetically inactive, either because it contains no genes or because the genes it does contain need not be expressed in that cell at that time.
Characteristically, heterochromatin replicates later in the S-phase of the cell cycle as a result of the higher degree of chromosome condensation. Heterochromatin is found in all eukaryotic species near the centromeres, at telomeres and elsewhere in a species-specific manner. Two classes of heterochromatin can be distinguished.
i. Constitutive Heterochromatin:
It is always genetically inactive and permanently condensed in all types of cell of a species. It is found at homologous sites on chromosome pairs.” This type of heterochromatin contains highly repeated DNA sequences, called satellite DNA, which might have a structural role in chromosomes. Centromeric and telomeric heterochromatin are example of constitutive heterochromatin.
ii. Facultative Heterochromatin:
It is potential to become condensed to the hetero- chromatin state. It may contain genes that are made inactive when the chromatin becomes condensed. Barr bodies i.e., inactivated X chromosomes in female mammals are examples of facultative heterochromatin.
Proteins associated with the DNA in chromatin are of two types – histones and non-histones. The DNA is wrapped around a core of histone molecule, and the non-histones are somehow associated with that complex.
Histones are small basic proteins with a high content of basic amino acid arginine and lysine and having net positive charges that facilitate their binding to the negatively charged DNA. Five main types of histones are found in chromosomes, viz., H1, H2A, H2B, H3 and H4. The four main histones, H2A, H2B, H3 and H4 are very similar in different species, being among the most conserved known proteins.
For example, only two amino acid differences exist in the H4 proteins of cow and peas, and only one amino acid difference between sea urchin and calf thymus H3. The remarkable similarity in the amino acid sequence of histones among different organisms is a strong indicator that histones play some basic role in organising the DNA in the chromosomes of all eukaryotes.
H1 is not conserved between species and has tissue-specific forms. It is present only once per 200 base pairs of DNA and is involved in the maintenance of a higher-order folding of chromatin. There are a large number of non-histone proteins that are usually acidic and are likely to bind to positively charged histones in the chromatin.
The enzymes and proteins involved with replication, transcription and the regulation of gene expression are example of chromatin-associated non-histones.
In contrast to histone, the non-histone proteins differ markedly in number and type from one cell type to another within an organism, at different times in the same cell type and from organism to organism. High mobility group (HM6) proteins are best example of non-histone proteins so far studied.
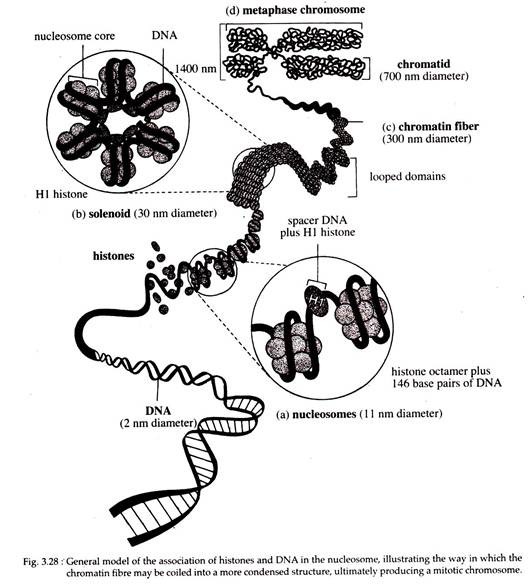
6. Molecular Organisation of Chromatin:
i. Nucleosomes:
The lowest level of chromosome organisation. Within the nucleus, the DNA double helix is coiled in a non-random way. There are several levels of packing of the DNA that allows a chromosome of several millimeters to even few centimeters long to fit into a nucleus that is only a few micrometer in diameter.
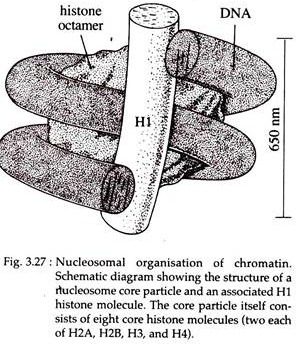
The simplest level of packing involves the winding of DNA around the histones in a spherical structure called a nucleosome.
These particles were discovered by Ada and Donald Olin’s and were initially referred to as u- bodies. These occur regularly along the axis of a chromatin strand. The more highly packed state involves higher order of coiling and looping of the DNA-histone complex, as seen in metaphase chromosomes. During gene expression, chromosome regions undergo changes from higher level of coiling to a lower level of coiling.
According to recent model, each nucleosome contains a core particle consisting of histone octamer, a single molecule of linker histone HI and super coiled DNA of about 146bp. This forms a chromatin subunit known as a chromatosome. In the histone octamer, two molecules of each of four core histones, H2A, H2B, H3, and H4 are present.
In test tube the core histones are capable of self-assembling into the histone octamer which is a flat-ended cylindrical particle, about 11 nm in diameter and 5.7 nm thickness.
The DNA appears to wrap (left handed) the outside of the nucleosome core about one and three-quarter times. The core particles are connected to one another by a stretch of linker DNA or spacer DNA of variable length, but typically comprising of about 60 base pairs.
Together, the DNA of nucleosome core and linker consist of about 200 base pairs. One molecule of H3 histone is normally located just outside the core particle. It is associated with both ends of the DNA as it enters and exits from the core particles (Fig. 3.27).
Electron microscopy reveals this DNA-protein complex as 10 nm chromatin fibre with an appearance of ‘beads on a string’, where the beads are the nucleosomes and “thread” connecting the ‘beads’ is naked linker DNA.
The more hydrophobic uncharged parts of the histones occupy the centre of the core, thereby promoting their aggregation into a tight core. In contrast, the polar basic parts of their proteins form flexible tails directed towards the outside of the particle where the positively charged residues are available to form ionic interactions with the negatively charged phosphates of the DNA backbone.
The paired sites on the surfaces of the histone octamer are thought to provide “docking pads” for the two strands of the DNA helix. Even though the DNA is tightly associated with the histone cote on its inner surface of the helical fibre, the outer surface of the DNA still remains exposed to make it accessible for interaction with other regulatory molecules.
Thus the formation of nucleosome represents the first level of packing, whereby the DNA helix is reduced to about one-third of its original length.
ii. Higher Level of Chromatin Organization:
In cell, chromatin typically does not exist in a “beads on a string” nucleofilaments; rather, it is kept in a more highly compact state. A general agreement, derived from electron microscopic observation, that the next level of packing above the nucleosome is the 30 nm chromatin fibre.
A number of models for the 30 nm fibre have been proposed on the basis of biophysical and biochemical data using both compact and relaxed chromatins. These studies reveal that histone HI plays an important role in the formation of the 30 nm fibres, since Hl- depleted chromatin can form 1 nm fibre, but not 30 nm fibres. However, the precise role of HI is currently unknown.
Coiling of the 10 nm nucleofilament to produce the 30 nm fibre creates a structure with 6 nucleosomes, per turn that was initially called a “solenoid” (Fig. 3.28b). However, this second level of packing, does not provide sufficient packing of the chromosome to explain the degree to which chromosomes are condensed during mitosis. So further folding of the chromatin must occur.
The 30 nm fibre forms a series of looped domains extending from a condensed protein lattice. The amount of DNA in each loop ranges from tens to hundreds of kilo-base pairs. The looped domains further condense the structure into chromatin fibre, which is 300 nm in diameter.
The fibres are then coiled into the chromosome arms that constitute a chromatid, which is a part of metaphase chromosome. Interphase chromosomes would be packed less tightly than metaphase chromosomes.
Since an interphase chromosome may have a diameter of 300 nm while a metaphase chromatid may have a diameter of 700 nm. It has been hypothesized that each loop is an independent unit of transcription and replication that functions independently of neighbouring loops. The diameter of chromatids varies among different organisms.
Recent studies by Timothy Richmond and his team (1997) on nucleosome crystals by X-ray diffraction revealed that there are several unstructured histone tails which are not packed into the folded histone domains within the core of the nucleosome. These tails help in the interaction with neighbouring structures.
The exact levels of packing beyond the 30 nm fibre are not clearly understood, till date. However, in the overall transition from the fully extended DNA helix to the extremely condensed status of a mitotic chromosome, a packing ratio of about 500 to 1 (the ratio of DNA length to the length of the structure that contains it) is achieved.
In fact, this model accounts for a ratio only of about 50 to 1. Obviously the larger fibre can be further bent, coiled, and packed during the formation of mitotic chromosome.
7. Special Types of Chromosomes:
The study of specialized chromosomes viz., polytene and lampbrush chromosomes reveals many insights into the arrangement and function of the genetic information. These chromosomes are so large that their organisation was discerned by light microscope long before the discovery of the mechanisms of formation of mitotic chromosomes from interphase chromatins.
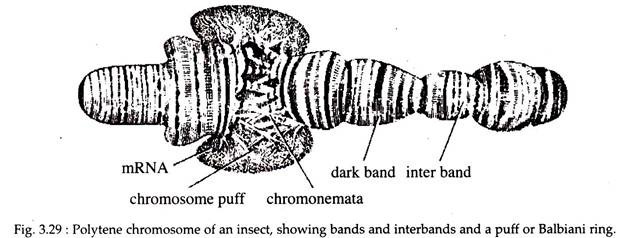
i. Polytene Chromosomes:
These type of chromosomes were first observed by F. G. Balbiani in 1881. Polytene chromosomes are large sized chromosomes with a linear series of alternating bands and inter-bands (Fig. 3.29). Individual bands are sometimes called chromomeres.
Each such chromosome is 200 to 600 (Jin long. The unusual appearance of these chromosomes results from several causes. First, these are paired homologues which are very unusual (somatic pairing). Second, in these chromosomes many DNA strands are present which undergo many rounds of replication but with no strand separation or cytokinesis (endomytosis).
So, as the replication proceeds, chromosomes are created having 1,000 to 5,000 DNA strands that remain in parallel register with one another. Distinctive band pattern along the long axis of the chromosome is due to the presence of such DNA strands. A band may contain up to 107 base pairs of DNA.
The strands present in the bands undergo localized uncoiling during genetic activity and forming puff-like structures (Fig. 3.29). These puffs, also called Balbiani rings, are active in RNA synthesis, as can be demonstrated by 3H-uridine labelling and radiography, in comparison to the bands that are not extended into puffs.
However, the relationship between the structure of the polytene chromosome and the genes contained within them is intriguing. These giant chromosomes are found in various tissues like salivary gland, midgut and Malpighian tubules, in the larvae of some flies (Drosophila), and in several species of protozoans and plants.
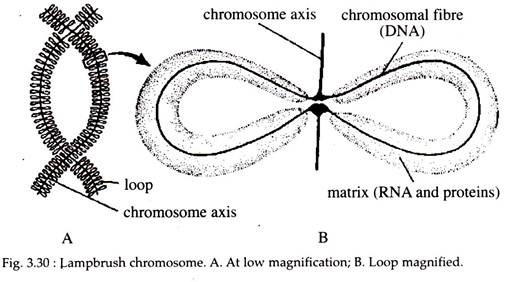
ii. Lampbrush Chromosomes:
In 1892, another specialized chromosome was discovered by Ruckert, in the oocytes of sharks, called the lampbrush chromosomes. It is so named because of its similarity to the brushes used to clean the chimneys of kerosene lamps.
It is very common in most vertebrate oocytes as well as the spermatocytes of some insects. Therefore, they are meiotic chromosomes found in prophase I. The lampbrush chromosomes are the bivalents in which the maternal and paternal chromosomes are held together by chiasmata at those sites where crossing-over has previously occurred.
However, instead of condensing, as most meiotic chromosomes do, lampbrush chromosomes are often extended to lengths of 500 to 800 µm. Later in meiosis, they revert to their normal length of 15-20 µm. So these chromosomes are interpreted as extended, uncoiled versions of the normal meiotic diplotene chromosomes.
The linear axis of each chromosome contains a large number of condensed areas, referred to generally as chromomeres and are made up of two DNA helices. Emanating from each chromomere is a pair of lateral loops, (Fig. 3.30) which give the chromosome its distinctive appearance.
There are about 10,000 loops per chromosome set. Each such loop is thought to be composed of one DNA double helix. These loops are the active sites for RNA synthesis. Thus, the loops are in a way similar to puffs in polytene chromosomes, representing DNA that has been reeled out from the central chromosome axis during transcription (Fig. 3.30).