In this article we will discuss about:- 1. Meaning of DNA Fingerprinting 2. Basis of DNA Fingerprinting 3. Applications.
Meaning of DNA Fingerprinting:
Presence or absence of restriction sites in the genome can be used as genetic markers. Similarly, another type of variation in the length of repetitive DNA sequence cluster may be used for a wide variety of genetic analysis. Variation may exist in the number of tandemly repeated DNA sequences present between two restriction enzyme sites.
These sequences are called minisatellites, which are clusters of nucleotides, from 2 to 100 nucleotides in length. For example, GGTTGGGTTGGGTTGGGTTG is composed of four tandem repeats of the five nucleotide sequence GGTTG. In human genome, such clusters are widely dispersed. Typically each repeat contains between 14 to 100 nucleotides, the number of repeat at each locus varies from 2 to more than 100.
These loci are called variable-number tandem repeats (VNTRs), because the number of repeats at a given locus is variable and each variation constitutes a VNTR allele. Many loci have dozens of alleles each, as a result, heterozygosity is produced.
ADVERTISEMENTS:
These polymorphisms in genome provide a unique molecular pattern for an individual and serve as the basis for a technique called DNA fingerprint or DNA typing or DNA profiling, developed by Alec Jeffreys and his colleagues at Leicester University, UK during 1985-86.
Basis of DNA Fingerprinting:
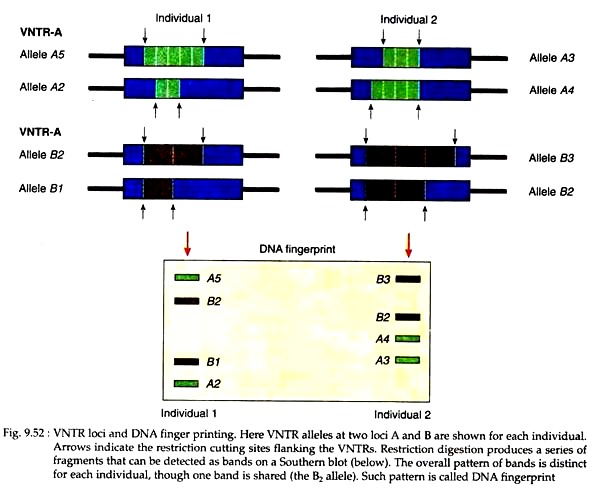
When VNTR sequences are cut with restriction enzymes and visualised by Southern blotting, the pattern of bands are called DNA fingerprint (Fig. 9.52). Because of differences in the number of repeats at each locus, the overall pattern of bands is distinct for each individual, but the pattern varies from individual to individual.
In fact, there is so much variation in band pattern from individual to individual that theoretically each person’s pattern is unique. Therefore, it represents a characteristic “snapshot” of the genome of an individual, allowing it to be distinguished from those of other individuals in the population.
ADVERTISEMENTS:
In the fingerprinting technique, first DNA will be isolated from the sample. It can be performed on very small quantities of materials (less than 60 microliters of blood) that may be present in various samples like blood, semen, hair roots, tears, saliva, perspiration etc. Very old DNA sample can also be used for this test. Following isolation, the DNA will be subjected to Southern blotting and DNA hybridisation with the help of specific DNA probes.
At present about a dozen of different VNTR probes are used in standard fingerprinting tests. The probes correspond to hyper variable minisatellites in DNA, each made up of tandem repeats of short sequences. Most of these are on different chromosomes and each probe can be used to visualize the VNTR pattern at a particular locus. To obtain a detailed DNA fingerprinting profile, four to ten different VNTR probes are used.
The results of DNA fingerprints are analyzed and interpreted with the help of statistics, probability and population genetics. In this way, the frequency of each VNTR allele in the population is calculated, and these are multiplied together to give a combined frequency, which is a calculation of how often the observed DNA fingerprint might occur in the population.
It is found that the probability of two individuals at random, that will produce the identical fingerprints of DNA bands is so low that it will never be encountered in such experiments, Therefore, if the DNA sample from the unknown individual from a site and the DNA sample from a suspected individual identically match with each other in terms of the fingerprint, then the identity is established between these two individuals. This scientific basis is accepted in the court of law.
Applications of DNA Fingerprinting:
ADVERTISEMENTS:
Since its discovery, DNA fingerprints have been used as evidences in the identification of criminals in murder case, rapists in rape case, parents in case of doubtful parentage etc. This method has also been used in a wide range of other applications, including immigration cases, disputes involving purebred dogs, and in animal conservation studies.
In Forensic Analysis:
If a DNA profile from tissue found at a crime scene matches with that of a suspect, it does not prove that the tissue belongs to the suspect; instead, it excludes all those who have a different DNA profile. Therefore, to generate five to more DNA profiles from the same sample, different probes have to be used.
The more profiles that match, between the sample and the suspect, the more unlikely it is that the sample at the crime scene came from someone other than the suspect. Sometimes DNA fingerprinting in criminal case may not involve the suspect’s own DNA, but rather the DNA of plants or animals that are closely present at the crime scene.
In Paternity case:
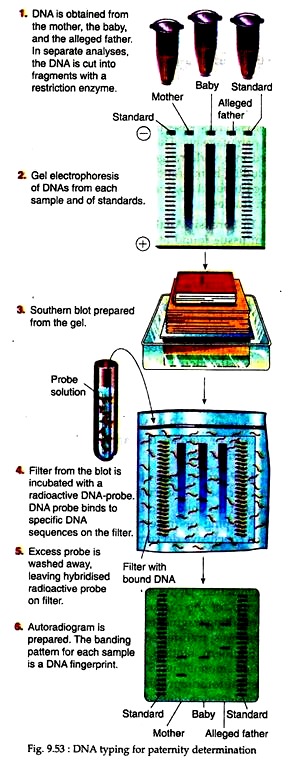
If a mother of a newborn accuses a particular man of being the father of her child, and the man denies it, the DNA typing or fingerprinting can at least reach conclusive decisions. In this case, DNA samples are taken from blood samples of mother, baby and the suspected father.
DNAs are cut with restriction enzyme for the marker to be analysed and the resulting fragments are separated by electrophoresis, transferred to a membrane filter by Southern blotting and probed with a labeled mono-locus STR or VNTR probe.
After autoradiography or chemiluminescence detection, the DNA banding pattern – the DNA fingerprint or DNA profile is analyzed to compare the samples (Fig. 9.53). The data shown can be interpreted as follows. In second lane two DNA fragments are shown from mother’s sample.
Therefore, the mother is heterozygous for one particular pair of alleles at the STR or VNTR locus under study. Similarly, two DNA fragments are also detected in the third lane for the baby.
It indicates that the baby is also heterozygous. It is also shown that one fragment for the baby matches the larger fragment of the mother’s DNA. The other fragment of the baby’s DNA is much larger, indicating presence of more repeats in that allele. In the autoradiogram it is also shown that the larger fragment of the baby’s DNA matches with one DNA fragment of the alleged father.
We know that the baby receives one allele from its mother and one from its father. The present data indicate that the man shares an allele with the baby, but he denied to be its father. If the man had no alleles in common with the baby, then the data would have proved that he is not the father. This is called exclusion result. The inclusion result that indicates positive identity is difficult to establish through DNA fingerprinting.
It depends on frequencies of STR or VNTR alleles identified by the probe in the ethnic population from which the accused person comes. However, for better’ confirmation, it is suggested to probe the DNA fragment with different mono-locus STRs or VNTRs and if the data for each probe indicated that the same person contributed a particular allele to the child, then the conclusion regarding the paternity will be more confirmed.
Other Applications of DNA Fingerprinting:
(i) Detection of genetically modified organisms (GMOs):
Genetically modified crops typically contain certain genes that were introduced for the development of new crops. In general, these genes express a particular promoter and a particular transcription terminator, enabling PCR primers to be designed on the basis of these sequences. Such primers can be used to test their presence.
A positive result will indicate that the plant is genetically modified or that the food contains one or many GMOs. On the other hand a negative result does not rule out the presence of a GMO, because the plant may have been genetically modified using genes with a different promoter or terminator.
(ii) Phylogenetic relationship:
Analysis and comparative study of DNA extracted from very ancient organisms, such as 40 million year old fossil leaf or 20 million year old insect in amber, or 40,000 year old mammoth with present day related organisms may be helpful to determine the actual phylogenetic relationship among present-day descendant.
(iii) Determination of variability in population or ethnic groups may be done by DNA typing.
(iv) In horses, dogs etc. pedigree status can be determined for certain breeds to prepare their breed registration.
(v) Conservation biology may also use DNA typing to determine genetic variability among endangered species.
(vi) Presence of pathogenic strains of bacteria like E. coli in food can be tested by DNA typing.