In this article we will discuss about the crossing over of genes, explained with the help of suitable diagrams.
The genes remain in linear order along the length of the chromosome and linkage is the physical relationship between the genes. During meiosis a physical crossover between gene pairs (Hi homologous chromosomes occur.
According to Jonssen’s crossover Theory (1909) a cytologically observed chiasma is the exchange point between the homologous chromosomes. Crossing over means breaks of linkage within the chromosome and physical exchange of gene from one chromosome to the corresponding position of the homologous one.
The crossing over may be in one, two, three or more points and are said to be single, double, triple or multiple crossing over and the gametes are called crossovers.
ADVERTISEMENTS:
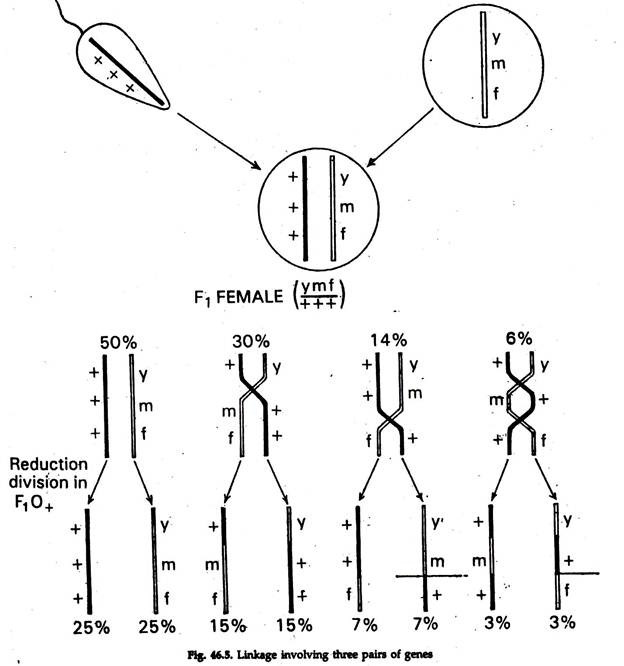
The experiment (Fig. 46.5) to show the example of linkage in a single crossover. The percentage of crossover varies between different genes but remains constant for each pair. The crossover percentage is dependent on the relative distance of the two pairs of alleles on the chromosome. Greater the distance, higher will be the amount of crossing over percentage.
It is found that crossing over in one region apparently inhibits or interferes with crossing over in a neighbouring region. Muller termed this as ‘interference’. There are only few or double crossovers within a 10 unit or less long section of chromosome due to interference. When the distance between two genes increases, the interference becomes less or even nil.
The double crossover is the coincidence or coming together of two single crossovers and involves three genes on the same chromosome. When double crossovers occur in expected numbers, the coincidence is considered as 100 per cent and interference is 0.
ADVERTISEMENTS:
Alternatively, when there is no double crossover, the interference is 100% and coincidence is 0. The ratio of observed frequency to the expected frequency of double crossovers is known as “coincidence coefficient”.
Double Crossover in Drosophila:
In Drosophila, yellow body (y), miniature wing (m) and forked bristles (f) are three recessive mutations in the X-chromosome. The normal fly possesses grey body (y+), long wings (m+) and straight bristles (f+). A cross between yellow-miniature-forked female ![]() and a normal male
and a normal male ![]() produces in f1 female with genotype
produces in f1 female with genotype ![]() During ovulation in the female, the chromosome might pair in four possible ways, resulting eight classes of combination (Fig. 46.5). The first and 2nd classes of combination
During ovulation in the female, the chromosome might pair in four possible ways, resulting eight classes of combination (Fig. 46.5). The first and 2nd classes of combination ![]() are the crosser over, the third and forth classed
are the crosser over, the third and forth classed ![]() are single crossovers between y and m, the 5th and the 6th classes
are single crossovers between y and m, the 5th and the 6th classes ![]() are single crossover between m and f and 7th and 8th classes
are single crossover between m and f and 7th and 8th classes ![]() are double crossovers between y and f. if these are test crossed with triple recessive male (y m f), then all these eight classes of offspring will be produced.
are double crossovers between y and f. if these are test crossed with triple recessive male (y m f), then all these eight classes of offspring will be produced.
Calculation of distance and order between y m f:
ADVERTISEMENTS:
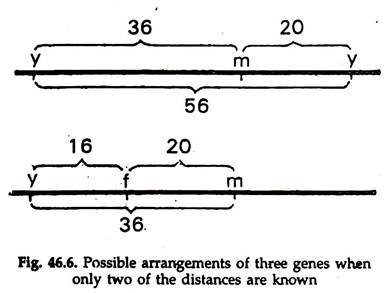
The distances between y m f can be calculated now by knowing the percentage of crossover. Ignoring the forked locus, the crossovers between y and m result in the combinations y+ m and y m+. These combinations are found in Fig. 46.6 as classes 3 and 4 (the single crossovers in between y m) and classes 7 and 8 (the double crossover between y f).
Percentage of single crossover between y and m is 30 and percentage of double crossover between y and m is 6. So, the total crossover between y and m is 30 + 6 = 36%. Similarly, the single crossover percentage between m and f is 14% and the double crossover is 6%. So, the total crossover percentage between m and f is 14 + 6 = 20.
Therefore, the distance between y and m is 36 and distance between m and f is 20.
We know the genes are in the order of y m f and the distance between y and f is 36 + 20 = 56 (Fig. 46.6). The double crossovers are counted twice, because a double crossover is equivalent to two single crossovers, one between y and m and another between m and f.
With the test cross the actual percentage of crossovers between y and m and f will give the real order of y m f.