The following points highlight the two main types of transposable elements found in prokaryotes. The types are: 1. Insertion Sequences 2. Transposon.
Type # 1. Insertion Sequences (IS):
IS elements are the simplest transposable elements and are normal constituents of bacterial chromosomes and plasmids. The IS elements range in size from 768 bp to more than 5,000 bp and are found in most cells. All such elements end with perfect or nearly perfect terminal inverted repeats (IR’s) of 9 to 41 bp.
Thus, essentially the same sequence is found at each end of an IS, but in opposite orientations. The IS elements encode an enzyme transposase that is essential for their own transposition. This enzyme recognises those sequences that initiate transposition.
Integration of IS elements along the chromosome may cause mutation by disrupting either the coding sequence of a gene or a gene’s regulatory sequence. In addition, the presence of IS elements can cause deletion or inversion type of mutations in the adjacent DNA. Finally, deletion and inversion events may also occur as a result of crossing over between duplicated IS elements in the genome.
ADVERTISEMENTS:
Example:
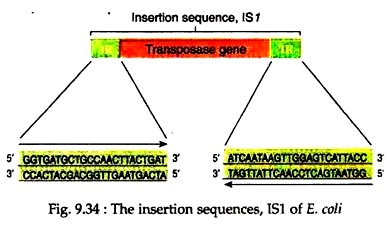
In E. coli, a number of IS elements are known, e.g., IS1, IS2, IS1OR etc. Each genome may contain up to 30 copies of IS elements and each has a characteristic length and nucleotide sequence. For example, IS1 has 768 bps and is present in 4 to 19 copies in E. coli chromosome. The inverted repeats of IS1 consist of 23 bp with not quite identical sequences (Fig. 9.34).
ADVERTISEMENTS:
Mechanism of transposition:
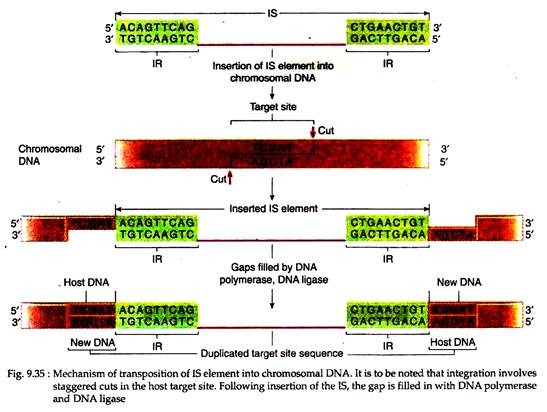
The IS elements integrate at a target site with which the element has no sequence homology. At the time of insertion, first a staggered cut is made at the target site and the IS element is then inserted, becoming joined to the single-stranded ends.
DNA polymerase and ligase then fill in the gaps, making an integrated IS element with two direct repeats of the target site sequence flanking the IS element. Here ‘direct’ means that the two sequences are repeated in the same orientation (Fig. 9.35). These small direct repeats are called target site duplications and their size varies from 4 to 13 bp.
Type # 2. Transposon (Tn):
The transposon is more complex mobile genetic element than the IS element. The Tn contains gene coding for transposase as well as other proteins.
ADVERTISEMENTS:
In prokaryotes, two types of transposons are found:
(a) Composite transposons and
(b) Non-composite transposons.
(a) Composite transposons:
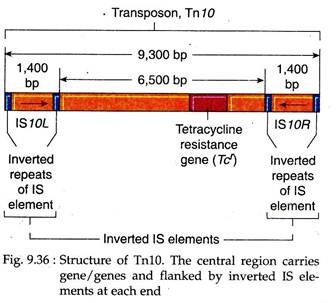
These Tn elements may be 1000 bp long and have a complex structure with a central region containing genes that confer resistance to antibiotics. They are flanked by IS elements of same type on both sides which are called ISL for the left one and ISR for the right one.
The ISs themselves have terminal inverted repeats in addition to terminal inverted repeats of the composite transposon. So the ISL and ISR may be in the same or inverted orientation, relative to each other.
Example:
ADVERTISEMENTS:
Tn10 (Fig. 9.36). Here the IS elements help in the transposition of the composite transposons. The transposase is supplied by one or both IS elements and recognise the inverted repeats of the IS elements at the two ends of the transposon to initiate transposition. Like IS elements, they produce target site duplication after transposition, which is 9 bp long in case of Tn10. Transposition of Tn10 is rare, occurring once in 107 cell generations.
(b) Non-composite transposons:
Unlike the composite transposon, non-composite transposons do not contain IS elements at their ends, but has the repeated sequences at their ends that are required for transposition.
Example:
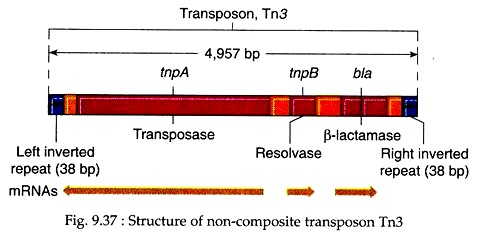
Tn3 (Fig. 9.37). It has 38 bp inverted terminal repeats and contains three genes in its central region. One of the genes is bla encoding β-lactamase which breaks down ampicillin and, therefore, makes the cells with Tn3 resistant to ampicillin.
The other two genes are tnpA and tnpB encoding the enzymes transposase and resolvase, respectively. Transposase catalyses insertion of Tn into new sites, and resolvase helps in the recombinational events associated with transposition. Like composite transposons they also cause target site duplication while they move. For example, Tn3 produces a 5 bp site duplication.
Mechanism of transposition:
Several models have been proposed regarding the transposition of transposons.
Two most popular models are discussed here:
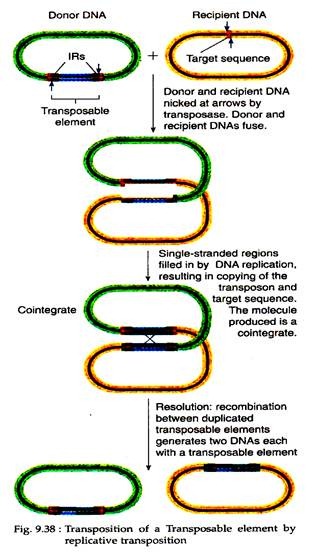
(i) Replicative transposition:
According to this model, transposition of transposon occurs from one genome to another. For example, from a plasmid to a bacterial chromosome or vice versa. In this case, the donor DNA with the Tn elements fuses with the recipient DNA. During this, the Tn element becomes duplicated with one copy located at each junction between donor and recipient DNA.
This fused product is called a co-integrate. Then, the co-integrate is resolved into two products, each with one copy of the transposable element. Since the Tn element becomes duplicated, this process is called replicative transposition (Fig. 9.38).
It involves two enzymes:
(a) Transposase acts on the end of original Tn element and
(b) Resolvase acts on the duplicated copies.
Tn3 and other non-composite transposons show this type of transposition.
(ii) Non-replicative transposition:
It allows the transposon to move physically from one location to another on the same or different DNA, without any replication of the element. This mechanism is called conservative transposition because here every nucleotide bond is conserved.
The Tn element is lost from the original position leaving a break at the donor DNA which is lethal unless it is repaired. Tn10 transposon transposes by conservative transposition. Insertion of a transposon into the reading frame of a gene may cause loss of function of that gene.