Contents:
- Meaning of Transcription
- Ingredients of Transcription
- Transcription of Protein Coding Gene in E. coli
- Initiation of Transcription: Template Binding
- Transcriptions of Other Genes; Prokaryotic rRNA Transcription
1. Meaning of Transcription:
The process by which RNA molecules are synthesized on a DNA template is called transcription. Genetic code for the protein’s amino acid sequence must be read from the DNA. RNA is the intermediate molecule in the process of information flow between DNA and protein.
Therefore, the significance of transcription is enormous as it is the initial step in the process of information flow within the cell. Genome of any organism consists of specific sequences of base pairs distributed in the chromosomes and genes include sequence of base pairs that are transcribed. Thus, the transcription process is also referred to as gene expression.
During transcription, generally only one of the two DNA strands is transcribed into an RNA. Four different types of RNA molecules, viz., messenger RNA (mRNA), transfer RNA (tRNA), ribosomal RNA (rRNA) and small nuclear RNA (snRNA) are produced by transcription.
ADVERTISEMENTS:
In prokaryotes, first three kinds of RNAs are found and only the mRNA molecule is translated to produce a protein. The gene that codes for an mRNA molecule and hence for a protein, is called structural gene. The genes that code for other RNA molecules are different from the structural genes because their RNA transcripts are the final products of gene expression, i.e., these RNAs function directly as RNA molecules.
2. Ingredients of Transcription:
1. DNA template.
2. Ribonucleoside triphosphates viz., ATP, GTP, CTP and UTP (NTPs).
3. RNA polymerase.
ADVERTISEMENTS:
4. Mg++, Zn++ etc.
RNA polymerase in E. coli:
The principal enzyme responsible for RNA synthesis is RNA polymerase which catalyzes the polymerization of ribonucleoside 5′ triphosphates. Unlike DNA polymerase, RNA polymerase does not require any preformed primer to initiate the synthesis of RNA.
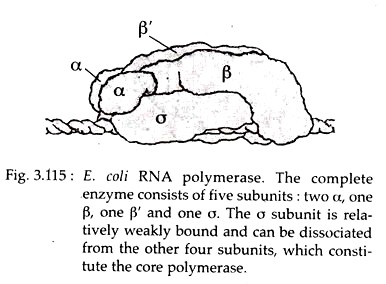
In E. coli, a single type of RNA polymerase catalyzes the synthesis of all type of RNAs. The active form of enzyme or holoenzyme contains the subunits α2ββσ and has a molecular weight of almost 5,00,000 Daltons (Fig. 3.115).
ADVERTISEMENTS:
In E. coli there may be more than one sigma (σ) factors, each associates with the same core enzyme at different times for expression of different genes. The σ is relatively weakly bound and can be separated from the core polymerase.
The latter is fully capable of catalyzing the polymerization of NTPs (nucleoside triphosphates) into RNA, indicating that σ is not essential for the basic catalytic activity. However, without a, the core polymerase initiates RNA synthesis in a random manner. Therefore, the σ subunit is required to identify the correct sites for transcription initiation.
3. Transcription of Protein Coding Gene in E. coli:
A prokaryotic protein coding gene has three sequences with respect to its transcription.
(1) The sequence up stream of the start of the coding sequence is called promoter with which RNA polymerase interacts.
(2) The coding sequence, the sequence of DNA base pairs transcribed by RNA polymerase into single-stranded mRNA transcript.
(3) The termination sequence, a sequence downstream of the end of the coding sequence that specifies where transcription will stop.
During transcription, RNA is synthesized in the 5′ → 3′ direction and only one of the two DNA strands is ready to make the RNA strand’, which is called the template strand. The 5′ → 3′ DNA strand complementary to the template strand and that has the same polarity as the resulting RNA is called non-template strand.
ADVERTISEMENTS:
4. Initiation of Transcription: Template Binding:
The initial step of transcription is referred to as template binding. In E. coli, initial binding is established when the RNA polymerase sigma (σ) subunit recognizes the promoter sequence. In E. coli, two consensus sequences generally are found in the promotor at -35 and -10 i.e., centered at 35 and 10 base pairs upstream from the coding sequences.
Sequences that are similar in different genes of the same organism or in one or more genes of related organisms are called consensus sequences. One 5’TATAAT3′ is located at -10 upstream (the -35 region) and called pribnow box; the other 5TTGACA3′ is located 35 nucleotides upstream (the -35 region).
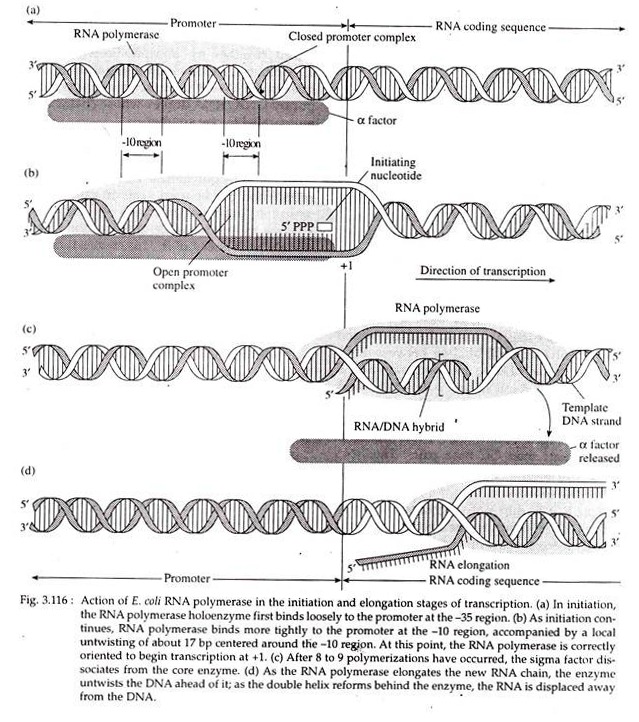
Following recognition of the promoter sequence by the sigma factor, the RNA polymerase holoenzyme binds to the promoter in two distinct steps. First, it binds loosely to the -35 region of the promoter while the DNA is still in double helical form (Fig. 3.116).
Second step involves a tight binding between RNA polymerase and DNA; it accompanies a local untwisting of about 17 base pairs of the DNA centered around the -10 region. This -10 region consists of all AT base pairs, having two hydrogen bonds and so are easier to break apart than GC base pairs. The RNA polymerase is then correctly oriented to start transcription.
In E. coli, different promoters may exist which differ slightly in their actual sequence. In fact there are several sigma factors (e.g., σ 70, σ28, σ 38) in E. coli which following binding to the core RNA polymerase, permit the holoenzyme to recognize different promoters. Most promoters have the recognition sequence as discussed above.
The sigma factor with a molecular weight of 70,000 Dalton, called σ 70 recognizes the above promoter. Another sigma called σ 32 can recognize the promoter with two different recognition sequences like CCCCC at -39, and TATAAA- TA at -16.
Elongation of RNA:
Following initiation i.e., recognition and binding of the RNA polymerase to the promoter, the enzyme catalyzes the insertion of the first 5′-ribonucleoside triphosphate which is complementary to the first nucleotide.
Subsequent nucleotide components are inserted and linked together by phosphodiester bonds. This process, called chain elongation continues in the 5′ → 3′ direction, creating a temporary DNA/RNA duplex whose chains runs antiparallel to one another.
Once 8 or 9 RNA nucleotides have been linked together, the sigma factor dissociates from the RNA polymerase core enzyme (Fig. 3.116a) and can be used again in other transcription initiation reaction.
As the core enzyme moves along, it untwists the DNA double helix ahead of it. This creates torsional stress in the DNA, so the DNA helix reform behind the enzyme as the latter moves along the template. In E. coli, transcription proceeds at the rate of about 30-50 nucleotides/second at 37°C.
Termination:
Termination of transcription in E. coli is signaled by the termination sequence or terminators of the gene that is about 40 base pairs in length. In E. coli one important protein called rho (p) factor is involved in the termination of transcription in some genes and is referred as rho-dependent terminators (type II terminators).
At many other terminators, the core RNA polymerase itself can cause termination, this process is called rho-independent terminators (type I terminators).
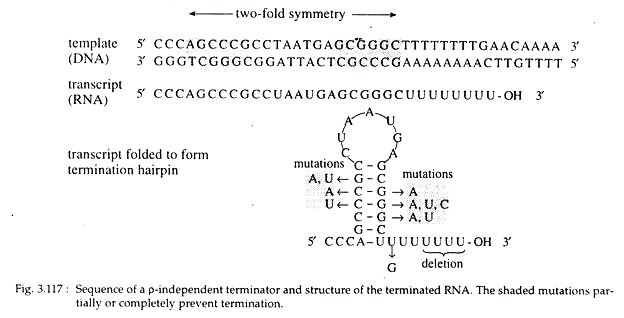
In the latter case, a sequence of two fold symmetry that are about 16-20 base pairs upstream of the termination point is present. A two fold symmetry is one that is approximately self-complementary about its centre; and half of the sequence is complementary to the other half.
The transcript of this region can form a hairpin loop (Fig. 3.117) which is followed by a string of about 4-8 AT base pairs giving rise to a series of Us in the RNA transcript which is just upstream of the transcription termination point.
This combination of hairpin loop and series of Us in the RNA leads to termination. It is assumed that rapid formation of hairpin loop destabilizes the RNA-DNA hybrid in the terminator region, which in turn causes the release of the RNA as well as transcription termination.
In case of p-dependent termination, no hairpin loop or string of us are found. The rho protein has two domains. One domain binds to RNA and the other domain binds to ATP. For termination, rho first binds to ATP and is activated, then it binds to recognition sequence in the terminator region when that region of the RNA transcript is synthesized.
The RNA transcript then somehow is unwound from the DNA template. Transcription then stops and both the transcript and RNA polymerase are released from the DNA template.
In prokaryotes, the RNA thus transcribed functions directly as mRNA molecules for translation. Moreover, since prokaryotes lack a nucleus, the mRNA begins to be translated on ribosomes before it has been completely transcribed.
This process is called coupled transcription and translation. Here, an exact point-by-point relationship exists between the order of the base pairs in the gene and the order of the corresponding bases in mature mRNA. This is referred to as co-linearity .between a gene and its primary transcript.
5. Transcriptions of Other Genes; Prokaryotic rRNA Transcription:
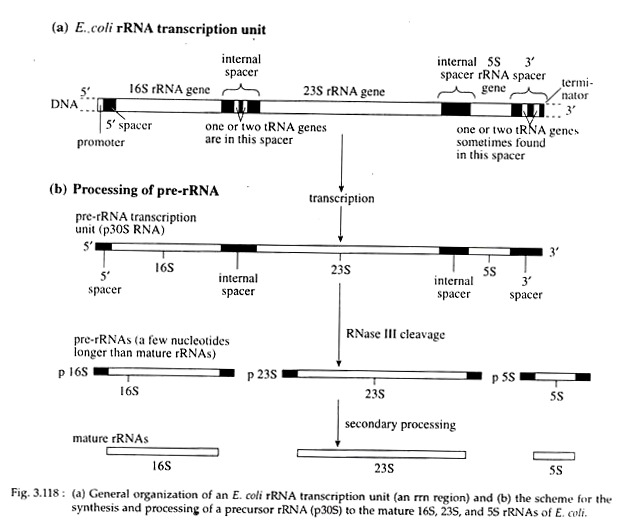
The regions of DNA that contain the genes for rRNA are called ribosomal DNA. In E. coli seven rRNA transcription units, each consists of one gene for the three rRNAs are present on the chromosome. In each transcription unit, the three rRNA genes are arranged in the order 16S — 23S — 5S (Fig. 3.118a).
However, one or two tRNA genes are also found in the internal spacer between 16S and 23S rRNA coding sequences. Another one or two tRNA genes are found in the 3′ spacer between the end of the 5S rRNA gene and the’ 3′ end in three of the seven transcription units. The tRNA genes are transcribed as part of the pre-rRNA molecule which are removed later on.
The overall transcription process is same as described for mRNA transcription. Here the transcription product is a 30S pre-rRNA which contains a 5′ leader sequence, the 16S, 23S, and 5S rRNA sequences (each separated by spacer sequences) and a 3′ trailer sequence (Fig. 3.118b).
As the rRNA genes are being transcribed, the pre-rRNA transcript rapidly becomes associated with ribosomal proteins. The mature 16S, 23S and 5S rRNAs are produced from their precursors by the action of some specific processing enzymes. The cleavage takes place within a complex formed between the rRNA transcript and ribosomal proteins.
Transcription of tRNA:
In E. coli, a cluster of tRNA genes may be transcribed to produce a single RNA transcript containing a number of tRNA sequences. The general organization of the multi-tRNA-pre-tRNA molecule is 5′-leader — (tRNA-spacer)n– tRNA-trailer -3′.
In such cases, the leader, the trailer and the spacer sequences are removed by specific enzymes. The enzyme RNase P catalyzes the removal of the 5′-leader sequence, and RNase Q catalyzes the removal of the 3′ trailer sequence.
From the above discussion it appears that in addition to coding sequence, gene contains other sequences that are important for the regulation of transcription called gene regulatory elements. In E. coli, mRNA is transcribed as the functional molecule, while processing events are confined to the tRNAs and rRNAs.